Living organisms are the result of interactions between their genomes and environment. Genomes are not just linear information to be translated into three-dimensional information which emerges as the phenotypes, depending on the environment. They are much more complex. They are far more complicated digital circuits than humans have ever invented.
“Let me now comment on the question “what next”. Up to now we are working in the descriptive phase of molecular biology. But the real challenge will start when we enter the synthetic phase of research in our field. We will then devise new control elements and add these new modules to the existing genomes or build up wholly new genomes. This would be a field with the unlimited expansion potential and hardly any limitations to building “new better control circuits” and finally other “synthetic” organisms.”
Szybalski, Polish geneticist (1974)
Following the Nobel for medicine to Arber, Nathans and Smith for discovery of restriction endonucleases (1978), he wrote,
“The work on restriction nucleases not only permits us easily to construct recombinant DNA and to analyze individual genes, but also has led us into the new era of synthetic biology where new gene arrangements can be constructed and evaluated”.
How we rearrange these circuits to impart novel properties to existing biological systems or build novel biological systems from scratch comprise both the excitement and the challenge of synthetic biology.
A ground-breaking paper showed last year that life, which is totally different from that around us, is possible. Philipp Holliger’s team at the MRC Laboratory of Molecular Biology in Cambridge, UK developed six xeno-nucleic acids (XNAs) which can carry genetic information like the DNA.
The key challenge for the team was to synthesize enzymes that would copy a gene from a DNA to an XNA and another enzyme to copy it back into DNA. The finding put to rest the long speculation whether life necessarily needs DNA or XNA. As Holliger points out,
“I would be surprised if we find truly extraterrestrial life that was based on DNA and RNA. There might be an XNA world on a different planet”.
Biology in the Basement
It is a widely known fact that Microsoft, Apple and a few other IT giants were started by students in garages. A similar trend is evident in some domains of biology as an emerging movement by the name of do-it-yourself (DIY) biology. DIY biologists are mushrooming across the States and Europe as small but connected groups.
These groups turn used pressure cookers into autoclaves, refrigerators into incubators and order used lab apparatus and genetic fragments online. An article recently revealed how the bioterrorists could hack the President’s DNA. The author argued in favour of the possibility of converting personalized medicine into personalized weapon. He even claimed that the US government was covertly acquiring world leaders’ DNA while protecting Barack Obama’s.
What he failed to see was that personalized medicine warrants not a different drug for each individual, but a different drug for each genetic subset of the population. DIY biologists, though, don’t aim this high. They are content by simple experiments such as making their yogurt glow in the dark, like Meredith Patterson.
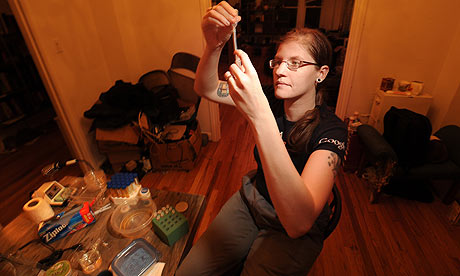
She isolated Lactobacillus acidophilus DNA from yogurt bought at a local grocery store. Her protocol for isolation of DNA involves addition of saline to the sample, adding some shampoo followed by meat tenderizer. After a while, she spun the mass in a salad spinner. She poured off the liquid and added some alcohol. Sticky strands of DNA were evident as white lump. [Disclaimer: You should not just read this but also try at home]. She ordered GFP (green fluorescent protein) gene from the Carolina Biological Supply Company. She transferred the gene into the bacteria through electroporation.
If people with kitchen labs and no formal experience can do such wonders, scientists with large laboratories can contribute even greatly. Synthetic biology may help address the global energy crisis. It’d be possible, in a decade or two, to design bacteria that produce biofuels out of organic material. Jay Keasling, a biochemical engineer, has developed a bacterium that rapidly pumps artemisinin, a key metabolite used in the treatment of malaria.
Who knows, the next big biotechnology company may come through some garage!
Open sourcing biological information
The major factor in the rapid growth in the computing sector over the last three decades or so has been open sourcing of information. Availability of program source codes online for free prompted young developers to come up with more efficient programs. Drew Endy believes a similar approach is feasible for biology too. Massachusetts Institute of Technology maintains a Registry of Standard Biological Parts which supplies DNA strings known to serve as a wide array of biological features. You can also register new DNA segments with novel functionalities. These standard biological parts are known as BioBricks.
It is hoped that the Registry will facilitate expansion of DIY biology (discussed more later) and advance the science quickly. In the same way that people can now design elegant websites (an example is the one you are currently on) without any knowledge of programming, in the future there might be do-it-yourself kits for gardeners to breed newer varieties, for people to screen pathogens in food or water samples and so on which could be used without any training of genetic engineering. Imagination is the limit!
MIT’s iGEM

Drew Endy transformed the student projects conducted during Independent Activities Periods in Massachusetts Institute of Technology into International Genetically Engineered Machine competition in 2003.
iGEM invites teams of undergraduates and high school students from all over the world to come up with biological systems with novel properties using the standard kits provided through the Registry. They can also come up with new BioBricks. Only five teams competed in 2004. But, now the competition has grown so popular that, since 2012, regional jamborees (Europe, Americas and Asia) are held in October, prior to the final jamboree in November.
Broader goals of the iGEM include enabling systematic engineering of biology, promoting open and transparent development of biological tools and involving society in application of biotechnology. The quality of projects that teams come up with in the iGEM competition spells much promise for the scope of synthetic biology.
These projects are not just-for-fun types but have immense commercial potential and social utility. Some of them are listed below –
- A cell-to-cell communication system that allows for the propagation of a set of instructions coded into a plasmid – TUDelft 2009
- Cost-effective red blood cell substitute constructed from engineered E. coli bacteria – UC Berkeley 2007
- Cancer specific RNA interference mediated cell destruction – Princeton 2007
- Designing a biological system to sense environmental glucose concentration and release insulin – Taipei 2007
- Solar powered bacterial fuel cell – Duke 2007
References
- Pinheiro, VB et al., Synthetic genetic polymers capable of heredity and evolution, Science, 20 April 2012
- Lego, R., Guess What’s Cooking in the Garage, Popular Science, 31 May 2012
- Official website of the iGEM Foundation

